Abstract
The Crenicichla mandelburgeri species complex from the Middle Paraná shows parallel evolution of ecomorphs to the unrelated C. missioneira species complex from the Uruguay River. In this article, we describe a new species from the C. mandelburgeri species complex that has evolved a parallel morphology and ecology to an unrelated species from the C. missioneira species complex (C. celidochilus). The new species is a pelagic predator that feeds predominantly on fishes and together with C. celidochilus is the only known pelagic species in the large riverine genus Crenicichla. The new species is endemic solely to a small tributary (the Urugua-í) of the Middle Paraná River where it is sympatric and partly syntopic with two other closely related endemic species that, however, differ strongly in their ecomorphologies (one is a generalistic invertivore and the other a specialized molluscivore). Mitochondrial DNA phylogeny finds the new species nested within the widespread C. mandelburgeri. Reduced genome-representation ddRAD analyses, however, demonstrate that this new species is of a hybrid origin and shares ancestry with C. ypo, one of the two studied sympatric species.
Similar content being viewed by others
Introduction
Cichlid fishes are one of the classical groups known for their repeated and spectacular adaptive radiations which make them excellent models in ecology and evolution (Fryer & Iles, 1972; Barlow, 2000; Seehausen, 2015). Adaptive radiation is an evolutionary response to ecological opportunity in which organisms diversify rapidly from an ancestral species into a multitude of new forms and where the rise of adaptations coincides with expansion of ecological diversity within a lineage (Simpson, 1953; Schluter, 2000; Givnish, 2015). The rapid accumulation of ecological roles and their associated adaptations is thus a hallmark of adaptive radiations (Simpson, 1953; Schluter, 2000). In African cichlids, the high diversity is associated with and often explained by the great amount of ecological opportunities present in lakes (Fryer & Iles, 1972; Joyce et al., 2005; Koblmüller et al., 2008), which may include access to new resources allowing for the exploration of novel phenotypes, or relaxed competition that may allow for exploration of novel regions of the adaptive landscape (Schluter, 2000). Lakes have thus served as the foundation for parallel adaptive radiations after their colonization by cichlid fishes (Seehausen, 2015). Series of lakes provide access to similar resources and adaptive landscapes, constrain immigration and emigration, and thereby trigger evolution of similar parallel ecomorphs despite independent evolutionary histories. In situ speciation is thus a feasible outcome in lakes and results in monophyletic groups that are endemic to that ecosystem (i.e., species flocks).
Rivers offer a lesser amount of niches than lakes, but despite this, riverine cichlid adaptive radiations and species flocks have also been proposed (Piálek et al., 2012; Říčan et al., 2016) and demonstrated among the Neotropical cichlids (Burress et al., 2018a, b). The clear disparity in commonness between lacustrine and riverine species flocks may be explained by a combination of factors (summarized from Seehausen, 2015) that usually distinguish lakes from rivers: (1) river communities are usually assembled by immigration such that disparate co-occurring ecomorphs are most likely from unrelated lineages that were united by active dispersal or by geodispersal (as opposed to speciation-assembled communities that arise in stable lakes), (2) many ecomorphs observed in lakes may be unlikely to evolve in rivers due to niches that are uncommon or temporally unstable in rivers, and (3) niches are partitioned to more minute ecological scales in lakes where stable conditions favor the evolution of accommodating processes such as resource partitioning.
Despite the relative paucity of in situ cichlid diversification in rivers (Říčan et al., 2016) and lack of cases of parallel evolution in rivers (Burress, 2015; Seehausen, 2015), one striking example of parallel evolution within rivers mirroring the situation in lakes has recently been found (Piálek et al., 2012; Burress et al., 2018b) in the Paraná and Uruguay Rivers of South America in pike cichlids (Crenicichla). These two Crenicichla assemblages (informally termed the C. mandelburgeri Kullander, 2009 species complex in the Paraná and the C. missioneira Lucena & Kullander, 1992 species complex in the Uruguay River) are each monophyletic and distantly related to one another, are endemic, and consist of sympatric species that co-occur throughout their drainage, and exhibit dramatic ecomorphological divergence within the two groups (Lucena & Kullander, 1992; Burress et al., 2013; Piálek et al., 2015) and parallelism between the two groups (Piálek et al., 2012, 2015; Burress et al., 2018b).
In this study, we describe a new species from the Paraná species complex and demonstrate that it is yet another parallel species to the Uruguay species flock. The new species is the first pelagic predatory species from the Paraná species complex. We use mtDNA and nDNA (ddRAD) phylogenies and morphological and habitat data to demonstrate the parallelism.
Materials and methods
The Unified Species Concept was employed in the present study (de Queiroz, 2007) in which species are equated with independently evolving metapopulation lineages. Our operational species-delimitation criteria were consistent morphological differences together with topology and divergence analyses in molecular phylogeny. Genetic sequences from the holotype specimen are included in the present study and follow the GenSeq nomenclature of Chakrabarty et al. (2013).
Morphological methods
Counts were taken as in Kullander (1986). Measurements were taken as described by Kullander (1986) for Crenicichla, and the pattern of their variation was visualized using principal component analysis (PCA) implemented in Canoco 5 (Microcomputer Power, Ithaca, NY, USA) according to Šmilauer & Lepš (2014). Measurements were taken as straight line distances with digital calliper to 0.1 mm on left side of specimen. Scale row nomenclature follows Kullander (1996). Body length is expressed as standard length (SL). Morphometric characters are expressed as percents of the SL. E1 scale counts refer to the scales in the row immediately above that containing the lower lateral line (Lucena & Kullander, 1992). Three paratypes were cleared and counterstained (C&S) following the method of Taylor & Van Dyke (1985). Pharyngeal teeth description and counts of frashed zone concavities follow Casciotta & Arratia (1993). In the descriptions, the number of specimens is indicated in parentheses; the holotype is indicated by an asterisk. Institutional abbreviations are as listed in Ferraris (2007).
Molecular methods
We have used the mitochondrial (mtDNA) cytochrome b (cytb) and NADH dehydrogenase subunit 2 (ND2) markers together with a reduced genome-representation nDNA dataset. The cytb and ND2 are the two well-resolving mitochondrial markers in Crenicichla (Piálek et al., 2012), and the ND2 marker even outperforms the much more widely used cytb marker in resolution and support on all hierarchical levels of Crenicichla phylogeny. The nDNA dataset was obtained using the double-digest Restriction-site Associated DNA Sequencing method (ddRADseq; Peterson et al., 2012). For both mtDNA and nDNA markers, we have sequenced a representative sampling of specimens covering the whole distribution of the C. mandelburgeri complex in Misiones, Argentina (Supplementary Table S1) plus several outgroup taxa from the remaining clades of the C. lacustris (Castelnau, 1855) group, and also representative samples of all main groups of Crenicichla (Piálek et al., 2012) which were needed for molecular clock dating analyses (only the closest outgroup species, C. vittata Heckel, 1840, is shown in Figs.).
Laboratory methods and datasets preparation
Genomic DNA was extracted from ethanol-preserved gill or fin tissue using the JETQUICK Tissue DNA Spin Kit (Genomed, Germany) following standard protocol. The primers and reaction conditions of polymerase chain reaction (PCR) amplification for the mtDNA loci are as described in Piálek et al. (2012). PCR reactions were performed in a Biometra TProfessional Cycler, and the PCR products were purified using the JETQUICK PCR Purification Spin Kit (Genomed). Sequencing was done by SEQme, Czech Republic (www.seqme.eu) and Macrogen Inc., Korea. Contiguous sequences of the gene segments were created by assembling DNA strands (forward and reverse) using GENEIOUS v. 11.0.2 (http://geneious.com; Kearse et al., 2012). Nucleotide coding sequences were also translated into protein sequences to check for possible stop codons or other ambiguities. Sequences were aligned using MUSCLE v. 3.8 (Edgar, 2004) with default settings.
The ddRADseq library was prepared as described in Říčan et al. (2016). Sequencing was performed on an Illumina HiSeq 2500 (1 + 0.4 lanes, 125 cycles P/E, v4 kit) in the EMBL Genomic Core Facility, Heidelberg, Germany. Basic characteristics of obtained reads were reviewed in FastQC v0.10.1 (Andrews, 2010). Barcode sorting and quality filtering of raw reads were performed in process_radtags (default setting), a component of Stacks v1.19 (Catchen et al., 2011). Retained RAD sequences were aligned onto the genome of the Neotropical cichlid species Amphilophus citrinellus (Günther, 1864) (GenBank GCA_000751415.1; https://www.ncbi.nlm.nih.gov). We used Bowtie 2 assembler v2.2.4 (Langmead & Salzberg, 2012) in ‘–very-sensitive-local’ mode, and the aligned tags were then processed in the ref_map pipeline as implemented in Stacks v1.35. Variant calling was processed in the population component of the Stacks pipeline. The resulting datasets either included only fixed (homozygous; masking thus within-individual polymorphism) or also all variable (heterozygous) loci.
All newly generated sequences were deposited in GenBank (Supplementary Table S1).
Phylogenetic methods
Bayesian inference (BI) analyses in BEAST v.1.8.4 (Drummond & Rambaut, 2007) were used for phylogenetic inference of the mtDNA data under substitution models inferred in jModelTest v0.1.1 (Posada, 2008) according to Akaike-corrected criterion (AICc). The BI analyses were run with partitioning into codon positions (1st + 2nd vs. 3rd) and using the Markov chain Monte Carlo (MCMC) simulation for 10 million generations with trees sampled and saved every 3000 generations. We used the relaxed molecular clock model with lognormal distribution of rates and for tree prior, the coalescent model with constant size. Four independent runs were performed to compare results of independent analyses. The analyses were run at the freely available Cipres server (https://www.phylo.org/). The first 10% of trees from each run were discarded as burn-in. Convergence of the runs was estimated with the use of TRACER v. 1.8.4 (Rambaut et al., 2014). The remaining trees from the four runs were used for reconstruction of the 50% majority-rule consensus tree with posterior probability (PP) values of the branches.
The ddRAD data were phylogenetically analyzed using maximum-likelihood criterion in RaxML v8.2.4 (Stamatakis, 2014). ML dataset was built in the denovo_map pipeline (minimum stacks depth, 7; minimum number of individuals with present locus, 70%) and included 49,952 variable SNPs. ML analysis was run with optimization of equilibrium frequencies and using the GTR + G substitution model as the most complex applicable one for the concatenated SNP matrix (Jones et al., 2013; Takahashi et al., 2014; Takahashi & Moreno, 2015), and 100 bootstrap replicates were used to evaluate statistical branch supports of ML trees.
Furthermore, we reconstructed species trees under the coalescent model using SVDquartets (SVDQ; Chifman & Kubatko, 2014). The SVDQ method does not rely on prior inference of individual gene trees, but it analyzes quartets of species in a coalescent framework using singular value decomposition of the matrix of site pattern frequencies and then assembles a species tree from the quartets using a supertree method. The SVDQ analysis was run as implemented in PAUP* v4.0b10 (Swofford, 2003) in the ‘species tree’ mode. SVDQ datasets were obtained from the reference-mapping pipeline ref_map (stacks depth, 7; minimum presence, 75%, fixed SNPs) and included 3,890 fixed sites. We sampled 1 million quartets assembled with QFM algorithm, and performed 100 bootstrap replicates of the data to assess branch support.
Molecular clock dating analysis in BEAST
For the calibration of the molecular clock, we have used an indirect secondary calibration based on the fossil-calibrated dated phylogeny of Musilová et al. (2015). The calibration date was set to 28 Ma with SD = 1 at the basal node of Crenicichla where it splits into the main Crenicichla species groups. The date estimates in the phylogeny of Musilová et al. (2015) are in very good agreement with the much larger all-cichlid study of Matschiner et al. (2017), which, however, could not be used directly for calibration within Crenicichla due to its limited sampling at below the genus level. A direct calibration also could not be used due to lack of Crenicichla fossils and since available Neotropical cichlid fossils are too old to use with only two mtDNA markers (Musilová et al., 2015). The BEAST analyses were run at the freely available Cipres server (https://www.phylo.org/). Runs were checked for convergence diagnostics with TRACER v.1.8.4 (Rambaut et al., 2014). Four well-converged runs were combined in LogCombiner v.1.8.4 with a burn-in of 10% for each of the data partition schemes. The final tree for each data partition scheme was produced from these data using TreeAnnotator v.1.8.4.
Admixture analysis
Population structure and individual ancestries for populations of closely related taxa to C. yjhui (C. mandelburgeri, C. ypo Casciotta, Almirón, Piálek, Gómez & Říčan, 2010, C. hu Piálek, Říčan, Casciotta & Almirón, 2010, C. sp. Paranay Guazú, C. sp. Piray Guazú, C. sp. Piray Guazú line) were analyzed using Admixture analysis. The analysis contained 121 individuals divided into 20 populations (defined by sampled drainages; Supplementary Table S1). The analyzed dataset obtained from the reference-mapping pipeline and based on variant calling in the populations program (Stacks v1.35; minimum stacks depth, 10; minimum number of populations with present locus, 60%; minimum number of individuals with present locus in each population, 60%) was LD-pruned in PLINK package v1.07 (Purcell et al., 2007; filtering with ‘–indep-pairwise 50 10 0.1′ setting) to obtain a subset of 15,066 variants in approximate linkage equilibrium (unlinked sites). To infer a population structure, the Admixture v1.3.0 program (Alexander et al., 2009) was run ten times with postulated number of ancestral populations K in the range of 2–10; optimal K was chosen based on the lowest cross-validation error (cv = 10).
Results
Taxonomy
Crenicichla yjhui Piálek, Casciotta, Almirón, Říčan, new species (Figs. 1, 2, 3, and 4; Table 1).
Crenicichla sp. ‘Urugua-í line’ Piálek et al. (2012) (Fig. 1, molecular phylogeny; Fig. 4, photo of live specimen).
Crenicichla sp. Urugua-í line Burress et al. (2018b) (Fig. 2, molecular phylogeny, morphological and trophic parallelism; Fig. 4, photo of preserved specimen and LPJ, trophic parallelism).
Holotype
MLP 11187, 131.9 mm, Argentina, Misiones, río Paraná basin, Embalse Urugua-í at camping (25°52′29.1″S, 54°33′02.2″W), April 2010, Casciotta et al. (Fig. 1).
Paratypes
All from Argentina, Misiones, río Paraná basin, arroyo Urugua-í basin. MLP 11184 (DNA A07-04-06), 1 ex. 96.8 mm, arroyo Falso Urugua-í (25°58′26″S, 54°15′29″W), September 2007. MLP 11183 (DNA116), 1 ex., 70.7 mm, arroyo Urugua-í (25°52′30″S, 54°33′00″W), April, 2010. MACN-ict 9555 (DNA 145–147, 149–152), 7 ex., 84.5–142.2 mm, Embalse Urugua–í at Policia Fluvial Station (25°54′02.2″S, 54°33′18.6″W), April 2010. MACN-ict 9556 (DNA 317), 2 ex., 83.5–95.7 mm, Embalse Urugua–í (25°52′30″S 54°33′00″W), April, 2010. MLP 11186, 17 ex. (3 c&s), 69.0–123,8 mm, Embalse Urugua–í (25°54′05″S, 54°33′19″W), December 2010. MLP11185, 19 ex., 72,8–97,5 mm, Embalse Urugua–í (25°54′05″S, 54°33′19″W), December 2010.
Diagnosis
Crenicichla yjhui belongs to the Crenicichla lacustris group (for distinguishing characters between the main Crenicichla species groups see Kullander, 1981, 1982, 1986) and within it to the C. mandelburgeri species complex (Piálek et al., 2012; Paraná species complex of Burress et al., 2018b).
Crenicichla yjhui is the only species in the C. lacustris group with body coloration composed solely of a conspicuous and continuous black lateral band. Crenicichla yjhui is further distinguished from all species of the C. mandelburgeri species complex (C. gillmorlisi Kullander & Lucena, 2013, C. hu, C. iguassuensis Haseman, 1911, C. mandelburgeri, C. taikyra Casciotta, Almirón, Aichino, Gómez, Piálek & Říčan, 2013, C. tapii Piálek, Dragová, Casciotta, Almirón & Říčan, 2015, C. tesay Casciotta & Almirón, 2009, C. tuca Piálek, Dragová, Casciotta, Almirón & O. Říčan, 2015, C. yaha Casciotta, Almirón & Gómez, 2006, C. ypo Casciotta, Almirón, Piálek, Gómez & Říčan, 2010) by the following combination of characters: absence in juveniles and adults of vertical bars along body, absence of dots or spots on flanks, and numerous scales (53–64) on E1 row (significantly overlapping only with the predatory C. iguassuensis and C. mandelburgeri, and the morphologically different herbivorous C. tapii).
Crenicichla yjhui is additionally distinguished from the most similar and most closely related species C. mandelburgeri and C. ypo by on average longer upper and lower jaws (ranges of these characters show overlap) and more delicate LPJ with longer lateral processes; from C. mandelburgeri additionally distinguished by the dorsal fin coloration that in most females has a wide black longitudinal band on the distal region bordered by white (vs. a single ocellus in C. mandelburgeri); and from C. ypo additionally distinguished by the absence of the wide longitudinal red stripe in the dorsal fin of females (vs. the presence in C. ypo).
Crenicichla yjhui is distinguished from C. vittata and C. jaguarensis Haseman, 1911 (phylogenetically closest and morphologically most similar species to the C. mandelburgeri complex) in the number of scales in a lateral row (53–64 vs. 78–85 in C. vittata and 47–53 in C. jaguarensis); from C. haroldoi Luengo & Britski, 1974 (Upper Paraná species close to the C. mandelburgeri complex) by the absence of dots on lateral line scales vs. the presence of brown dots on each lateral line scale; and from C. jupiaensis Britski & Luengo, 1968 (Upper Paraná species close to the C. mandelburgeri complex) by the presence of a well-developed suborbital stripe, scaled cheek, and the absence of a thin black line on the posterior margin of the preopercle vs. suborbital stripe reduced to a few spots posteriorly to the orbit, naked cheek, and the presence of a thin black line on the posterior margin of the preopercle.
Description
Body elongate, depth 4.6–5.5 times in SL (Fig. 1). Head as deep as wide or slightly deeper. Snout bluntly pointed in lateral view, 2.6–3.2 times in head length. Lower jaw slightly prognathous. Tip of maxilla reaching anterior margin of orbit in specimens over 80.0 mm SL. Lower lip folds widely separated along symphysis. Nostrils dorsolateral, nearer anterior margin of orbit (13) or equidistant between orbit and snout tip (11*). Posterior margin of preopercle serrated. Scales on flank strongly ctenoid. Head scales cycloid. Predorsal and prepelvic scales small. Interopercle naked. Cheek scaled, 6–9 scales below eye embedded in skin. Scales in E1 row 53 (1), 54 (3), 55 (3), 56 (3*), 57 (2), 58 (1), 59 (2), 60 (1), 61 (3), 62 (4), 64 (1). Scales in transverse row 10/13 (4*), 10/14 (2), 10/15 (11), 10/16 (2), 10/17 (1), 11/13 (1), 11/15 (1), 11/16 (2). One to three scale rows between lateral lines. Upper lateral line scales 16 (1), 21 (1), 23 (3), 24 (9), 25 (6), 26 (1), 28 (1), 29 (1*), 30 (1). Lower lateral line scales 6 (1), 10 (3*), 11 (7), 12 (9), 13 (4) plus 1–3 scales on caudal fin. Dorsal, anal, pectoral, and pelvic fins naked. Dorsal fin XX,12 (1); XXI,10 (1); XXI,11 (3); XXI,12 (4); XXII,10 (2*); XXII,11 (8); XXII,12 (2); XXIII,11 (3). Anal fin II,9 (1); III,8 (12*); III,9 (10); III,10 (1). Pectoral fin 15 (8*); 16 (12); 17 (4). Caudal-fin squamation not reaching the middle of fin. Soft-dorsal fin rounded or pointed tip, surpassing caudal-fin base. Tip of anal fin reaching or not caudal-fin base (reaching in holotype). Caudal fin rounded. Pectoral fin rounded, usually not reaching the tip of pelvic fin. Microbranchiospines present on second through fourth gill arches. Gill rakers externally on first gill arch: 3 on epibranchial, 1 on angle, and 8–9 on ceratobranchial (3 c&s). Three to five patches of unicuspid teeth on fourth ceratobranchial. Lower pharyngeal tooth plate with unicuspid recurved and bicuspid crenulated curved anteriorly teeth, those of posterior and medial row larger than remaining ones (Fig. 2). Upper pharyngeal tooth plate with unicuspid and bicuspid teeth. Frashed zone bearing one concavity with small unicuspid teeth. Premaxillary ascending process longer than dentigerous one. Premaxilla with 29–31 (3 c&s) unicuspid teeth on outer row, larger than inner ones. Five teeth rows near symphysis. Dentary with 29–33 (3 c&s) unicuspid teeth on outer row, 5 rows near symphysis. Total vertebrae: 36–38 (3 c&s). Premaxillary and dentary outer row teeth slightly movable, inner ones fully depressible.
Color in alcohol
Background of body pale gray or brown. Gray preorbital stripe between anterior margin of orbit and snout tip. Postorbital stripe between posterior margin of orbit and distal margin of preopercle black. Suborbital stripe black and variable in size, usually reaching the middle of cheek (holotype) or the anterior margin of preopercle; in some specimen restricted to the base and few dots. Suborbital stripe usually entire at base and fragmented distally with 3–7 dots wider (4 in holotype). Flanks with a black lateral band with indistinct margins dorsally reaching caudal-fin base; lateral band 4½ scales deep anteriorly, 2 scales deep posteriorly, lower margin running along lower lateral line tube row. Four or five wide vertical gray bars on dorsum below dorsal fin incompletely divided by a lighter zone; these bars can surpass below lateral band in adults (n = 9). Dorsal fin in females grayish, with long black horizontally extended band bordered with white between spines 14 or 15 to 6 or 7 branched ray; one specimen with a black subcircular spot ringed by white between spines 15–17. Dorsal fin in males also grayish with 2–3 irregular rows of dark spots on spinous portion and up to 4–8 on rear part on soft portion. Anal fin in females smoky with white lower margin; in males lower margin dark gray with 3–6 irregular rows of dark spots on posterior membranes. Pectoral fin pale gray. Pelvic fin whitish, dusked on anterior rays. Caudal fin dusky with 3–8 rows of dark dots, a black subcircular spot sometimes ringed with white, in some specimens black spot confluent with lateral band. Some specimens with membranes of two middle rays black, not reaching distal margin (Fig. 1).
Color in life
Background of middorsal body olive green to orange, mid ventral pale yellow. Preorbital and postorbital deep gray. Suborbital stripe black or deep brown. Flanks with black lateral band, yellow and orange pigment on flank at level and behind pectoral fin. Dorsal, anal, caudal, and pectoral fins pale olive green, pelvic pale yellow. Males with numerous dark scattered dots on dorsal, anal, and caudal fins. Caudal fin with a black subcircular spot bearing sometimes a white ring. Females with a distinctive coloration of the dorsal fin, with a wide black longitudinal stripe on the distal region of dorsal fin bordered by white (Figs. 3, 4).
Distribution
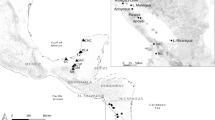
Crenicichla yjhui is found only in the arroyo Urugua–í basin, Paraná River basin, in Misiones Province, Argentina (Fig. 5). The arroyo Urugua–í (see Discussion) was originally isolated from the Paraná River by the 28 m high Urugua-í falls located eight km from the confluence with the Paraná River. In 1989, the falls were destroyed by the construction of the Urugua-í hydroelectric dam.
A Map of Middle Paraná basin with distribution of C. yjhui (red dots), the sympatric C. ypo (green dots) and C. mandelburgeri (blue dots). Dark blue and light blue dots show the northern and southern mtDNA clades (Fig. 6) of C. mandelburgeri, respectively. White bars show location of significant waterfalls. White and grey (in the Iguazú) dots show localities (distribution areas) of other species in the C. mandelburgeri complex. B Inset map of the Urugua-I basin. Red outline shows the extent of the artificial Urugua-í reservoir, star shows type locality of C. yjhui, and large and small dots show localities where the species is common and rare, respectively
Habitat
The arroyo Urugua–í basin has moderately fast flowing streams with an average depth of 1 m during the low water period outside of the dam influence. It bears macrophytes such as Echinodorus uruguayensis Arechavaleta, 1903 and Potamogeton pseudopolygonus Hagstrom, 1916. The bottom consists of bedrock, sand with gravel, and mud in the pools. After the dam construction some parts of the artificial lake are up to 6 m deep with relatively clear-water and the whole lake has a soft bottom (Supplementary Fig. S1). Some original localities of several species endemic to the basin (Australoheros tembe Casciotta, Gómez & Toresani, 1995, Crenicichla yaha, C. ypo, Gymnogeophagus che Casciotta, Gómez & Toresani, 2000 among the cichlids) from before the construction of the dam are now deep below the water surface. Crenicichla yjhui was not collected during the 1986 and 1987 studies before the hydroelectrical dam was built in 1989. This does not necessarily mean that the species was absent because the species is very rare even now outside the artificial lake area (only two specimens collected in 10 years, with just one specimen collected during the intensive campaign to rediscover C. yaha (Říčan et al., 2017; see below). It is only in the artificial lake where C. yjhui is common, with C. ypo being also reasonably common there (C. ypo is the dominant species in the streams, i.e., the area outside the lake influence). The third species C. yaha was only recently rediscovered and is very rare in the streams and absent from the artificial lake (Říčan et al., 2017). The pelagic morphology of C. yjhui (see Discussion) well reflects its preferred habitat of the artificial lake within the Urugua-í basin. Crenicichla yjhui is the rarest species of the three Crenicichla species in the streams of the Urugua-í basin.
Etymology
The specific epithet yjhui is composed of the Guaraní words Y (water) and Jhuí (arrow), meaning “water arrow.” A noun in apposition. The name is given in allusion to the streamlined head and body shape, coloration (a continuous black stripe resembling an arrow) and trophic ecology (open-water predator) of the species.
-
Genseq-1 CYTB. MLP 11187; GenBank accession number JF519974.
-
Genseq-1 ND2. MLP 11187; GenBank accession number JF520108.
Phylogeny
mtDNA
The mtDNA analysis (Fig. 6) reconstructs C. yjhui as a clade (without significant support; PP 0.8 in BI analysis), but the clade is contained within C. mandelburgeri (PP 1) between two clades that make up this nominal species.
MtDNA BEAST phylogeny of the C. mandelburgeri complex based on two mitochondrial markers (cytochrome b, ND2) with posterior probability (in squares) and molecular clock dates in Ma. Black dots show posterior probability of 1. Crenicichla yjhui, and its two candidate sister species based on nDNA ddRAD analyses (Figs. 7, 8; Supplementary Table S2) are colored. The Iguazú species and their clades are shown with gray branches. Blue branches show those Middle Paraná species that have had their mtDNA swept from the Iguazú, with C. mandelburgeri based on our interpretation (see Discussion) as the species that has spread this mtDNA throughout the Middle Paraná basin. The distributions of southern (S) and northern (N) clades of C. mandelburgeri are shown in Fig. 5
nDNA
In the nDNA genomic ddRAD analyses C. yjhui is always strongly monophyletic (BS 100) and depending on parameters (Supplementary Table S2) and method of analysis (Figs. 7, 8) is either found as the sister species of C. mandelburgeri or of C. ypo, always with 100% bootstrap support in all cases. Both ML and the species-tree SVDQ analyses thus support sister–species relationships of C. yjhui with both C. mandelburgeri and C. ypo.
SNP-based ddRAD maximum-likelihood (ML) phylogeny of the C. mandelburgeri complex. Node support is represented by bootstrap values (black dots show bootstrap of 100); numbers behind taxa show numbers of analyzed specimens. The Iguazú clade is shown with gray branches. Note that different analyses of the SNP data (Figs. 8, 9) place C. yjhui as a sister species of C. mandelburgeri or C. ypo with similar probabilities. The new species C. yjhui and its putatively closely related species (C. mandelburgeri, C. ypo) are highlighted by different colors
Species tree based on SVDquartets analysis using a 60% SNP matrix based on fixed (homozygotic) sites. See Supplementary Table S2 for results of analyses with 50% and 70% SNP matrices and variation of other parameters which find C. ypo and C. mandelburgeri as equally probable sister species of C. yjhui. The Iguazú clade is shown with gray branches. The new species C. yjhui and its putatively closely related species (C. mandelburgeri, C. ypo) are highlighted by different colors
The conflicting mtDNA phylogenetic position of C. yjhui is based on comparisons with the nDNA topologies (and based on Admixture analysis; see below) likely the result of introgression followed by sweep of mtDNA from the Iguazú clade into most Middle Paraná species including C. yjhui. Only C. hu and C. ypo have their original mtDNA based on this interpretation.
Molecular clock dating analysis in BEAST
Crenicichla yjhui separated from C. mandelburgeri based on mtDNA divergence age estimation (Fig. 6) around 0.41 Ma (95% HPD 0.20–0.56 Ma). Since the mtDNA phylogenetic position of C. yjhui is likely the result of an introgression and sweep of mtDNA the age estimation probably only shows the final separation of C. yjhui from C. mandelburgeri following the introgression. See Discussion for nDNA (ddRAD) divergence age estimation from Burress et al. (2018b).
Admixture analysis
The admixture analyses of the nDNA ddRAD mirror the results from the phylogenetic analyses of the same data. The K = 5 analysis finds C. yjhui as an admixed group composed approximately 50/50 from the allopatric C. mandelburgeri and the sympatric C. ypo, the optimal K = 6 and higher K value analyses support C. yjhui as a separate species (Fig. 9).
Inference of population structure based on the Admixture analysis; the optimal number of ancestral populations is marked with an asterisk. Note 50/50 assignment of C. yjhui between C. ypo and C. mandelburgeri at the suboptimal K5 and a separate species cluster at the optimal K6 and higher. Also note support for C. sp. Piray Guazú and C. sp. Paranay Guazú as separate groups
PCA of morphometric data
The PCA scatter plot indicates substantial separation of C. yjhui from the most similar C. mandelburgeri and complete separation from all remaining species including C. ypo. Crenicichla yjhui is found as the most streamlined species (lowest body depth, head depth, caudal peduncle depth, dorsal-fin height) with the longest jaws (Fig. 10). PC1 and PC2 explain 37.16% and 18.26% of the total variation, respectively.
Morphometric variation and discrimination of the described Middle Paraná species (except the Iguazú species and C. gillmorlisi) plus C. yjhui analyzed by PCA. Morphological measurements of 87 specimens (above 70 mm SL) measured following Kullander´s (1986) methodology for Crenicichla were taken as proportional values in % of SL
Discussion
Based on molecular phylogenies and on morphological characters C. yjhui forms an unambiguously diagnosable species that belongs to the C. mandelburgeri complex within the C. lacustris species group (Piálek et al., 2012; Burress et al., 2018b). The C. mandelburgeri group is endemic to the Middle Paraná River basin, including its main tributary, the Iguazú River. Within the C. mandelburgeri group the Iguazú River holds the highest diversity of sympatric and syntopic species (four; Piálek et al., 2015). The Urugua-í basin is with the discovery and description of C. yjhui the second richest tributary with three sympatric but only partially syntopic species (C. ypo with C. yaha in the streams, C. ypo with C. yjhui in the artificial lake).
Morphological differentiation of C. yjhui
Crenicichla yjhui (Figs. 1, 3, and 4) is most similar to C. mandelburgeri (Fig. 11) from which type populations (Kullander, 2009) it is easily distinguished in a number of characters that include a longer snout (10.6–12.3% SL vs. 6.7–9.8, mean 11.5 vs. 8.3–8.6), smaller orbital diameter (6.2–7.7 vs. 7.6–9.6, mean 6.8 vs. 8.2–8.5), smaller interorbital width (5.2–7.0 vs. 4.2–5.9; mean 6.1 vs. 4.8–5.0), longer upper (11.4–13.7 vs. 9.7–12.0, mean 12.5 vs. 10.8–11.0) and lower (15.3–17.1 vs. 13.5–15.8, mean 16.0 vs. 14.8–15.0) jaws, more scales in E1 row (53–64 vs. 44–56), a slightly narrower body (18.3–21.8 vs. 18.4–23.9, mean 19.5 vs. 20.6–21.5), a coloration composed chiefly of a continuous midlateral band, the dorsal fin of most females with a wide black longitudinal band on the distal region bordered by white (as in the sympatric and syntopic C. ypo; Fig. 12) unlike in C. mandelburgeri (Fig. 11) where females have a black ocellus (a rounded blotch bordered by white), and more delicate LPJ. The LPJ of C. yjhui has longer processes and smaller dentigerous area than C. mandelburgeri (Burress et al., 2018b; cf. Fig. 2 with Kullander, 2009), i.e., is more delicate as is typical for more piscivorous species (Burress et al., 2013, 2015; Burress, 2016; Burress et al., 2018a, b).
Differentiating C. yjhui from C. mandelburgeri sensu Kullander (2009) is thus straightforward in both qualitative (mainly coloration) and quantitative (morphometric and meristic) characters. Crenicichla mandelburgeri, however, has a much larger distribution than reported in Kullander (2009) and differentiating C. yjhui from the geographically closest (northern) populations of C. mandelburgeri is more difficult in quantitative characters. The northern part of the distribution of the nominal C. mandelburgeri contains a larger variation in morphology due to syntopy of C. mandelburgeri with other species (e.g., in the Piray Miní and the Piray Guazú; Fig. 5) and this area additionally contains several putative new species similar to C. mandelburgeri apart from C. yjhui (C. sp. Paranay Guazú, C. spp. Piray Guazú; Figs. 6, 7, 8, and 9; C. sp. Aguaray Guazú, C. sp. Ñacunday, C. sp. Yacui Guazú, C. sp. Monday; C. hu; Fig. 5). All quantitative characters show much higher overlap between C. yjhui and the northern C. mandelburgeri (upper jaw 11.4–13.7 vs. 10.7–13.2, mean 12.5 vs. 11.9; lower jaw 15.3–17.1 vs. 14.7–15.9, mean 16.0 vs. 15.6; body depth 18.3–21.8 vs. 19.3–24.6, mean 19.9 vs. 21.5), and in some there is no separation, and actually a reverse trend (snout 10.5–12.5 vs. 10.6–12.3% SL mean 11.6 vs. 11.5; orbital diameter 5.7–6.8 vs. 6.2–7.7, mean 6.2 vs. 6.8; interorbital width 5.9–7.6 vs. 5.2–7.0, mean 6.4 vs. 6.1; scales in E1 row 56–65, mean 60 vs. 53–64, mean 57.9). Multidimensional PCA analysis distinguishes C. yjhui and the northern C. mandelburgeri populations mainly based on the jaw lengths (Fig. 10).
Crenicichla yjhui is much more easily distinguished from its other closely related species C. ypo (Casciotta et al., 2010) in both qualitative (mainly coloration, in particular, the midlateral line and the absence of vertical bars and blotches vs. the absence of a midlateral line and the presence of vertical bars and blotches in C. ypo and then the red coloration of the base of the dorsal fin in adult females of C. ypo, cf. Figs. 3, 4 vs. 11) and quantitative characters (in C. yjhui a smaller interorbital width with 5.2–7.0 vs. 6.2–8.0, mean 6.1 vs. 7.1; longer upper jaw 11.4–13.7 vs. 11.1–13.0, mean 12.5 vs. 11.7, longer lower jaw 15.3–17.1 vs. 14.4–16.3, mean 16.0 vs. 15.4; narrower body 18.3–21.8 vs. 20.5–23.9, mean 19.9 vs. 22.3; more scales in E1 row 53–64, mean 57.9 vs. 47–55, mean 52.2 and a more delicate LPJ; Burress et al., 2018b; Fig. 10).
Ecomorphological parallelism of C. yjhui
Most characters that differentiate C. yjhui from C. mandelburgeri (longer jaws, narrower body, and more delicate LPJ) and C. ypo (longer jaws, more delicate LPJ, narrower head, narrower body; more scales along the body) including the coloration composed chiefly of the continuous midlateral band can be explained as associated with the increased levels of piscivory in C. yjhui and also open-water habitats (Seehausen et al., 1999; Burress et al., 2013, 2018a, b).
Based on our analyses of the morphology, habitats, and distribution of C. yjhui, we believe that we have presented a convincing case of a novel pelagic piscivorous pike cichlid, the first such known from the C. mandelburgeri species complex. This conclusion is also supported by the study of Burress et al. (2018b) who have analyzed the evolution of trophic traits (body and head shape, LPJ shape and mass) in the C. mandelburgeri complex (and the C. missioneira complex; see below) using landmark-based geometric morphometrics with focus on the two main evolutionary axes documented in cichlid radiations (the benthic-to-pelagic habitat axis and the soft-bodied to hard-shelled prey axis). Crenicichla yjhui, based on results of that study, is a strongly piscivorous species, together with C. iguassuensis and C. vittata (all other species in the group including C. mandelburgeri either have an ancestral morphology, or have evolved a different novel trophic state). Based on Burress et al. (2018b), C. yjhui has evolved a narrow trophic parallelism within the piscivorous ecomorph to C. celidochilus Casciotta, 1987 from the C. missioneira group. Both these species share the most upturned mouth and most delicate LPJ, streamlined body and coloration, uniquely shared among both the species groups composed of a continuous and dominant midlateral band (Burress et al., 2018b). Burress et al. (2013, 2015, 2018a) have demonstrated that these traits are in C. celidochilus associated with a pelagic hunting strategy that was additionally supported by pelagic fish prey dominating its stomach contents (as opposed to the other predatory species in the C. missioneira group where benthic prey items dominated).
Reticulate evolution of C. yjhui
Crenicichla yjhui appears to be a result of reticulate evolution based on our analyses of molecular markers. In the mtDNA markers used by Piálek et al. (2012) and in the here-extended phylogeny (Fig. 6) based on two mtDNA markers (cytb plus ND2), C. yjhui forms a clade, but the clade is contained within C. mandelburgeri, which forms two clades.
In nDNA genomic ddRAD analyses (Figs. 7, 8; Supplementary Table S2; Burress et al., 2018b), C. yjhui is depending on parameters of the analyses either found as sister species of C. mandelburgeri or of C. ypo. Admixture analyses of the nDNA ddRAD data mirror the results from the phylogenetic analyses of the same data. Low K value analyses find C. yjhui as an admixed group composed approximately 50/50 from C. mandelburgeri and C. ypo, and optimal and higher K value analyses find C. yjhui as a separate species-level cluster. This conflicting phylogenetic signal in nDNA is also resolved by TreeMix analyses (Piálek et al., 2018) which demonstrate shared ancestry with C. ypo and secondary contact and introgression from C. mandelburgeri.
Based on the mtDNA (cytb plus ND2) topology and divergence age estimation (Fig. 6), C. yjhui separated from C. mandelburgeri (i.e., following the secondary contact established above) around 0.41 Ma (95% HPD 0.20–0.56 Ma). Based on nDNA (ddRAD) divergence age estimation (Burress et al., 2018b) using a different and much younger calibration (2.6 times younger) the primary separation of C. yjhui from C. mandelburgeri/C. ypo occurred around 0.85 Ma. Using the same calibration as employed here for the mtDNA markers (see Methods) the nDNA divergence would equal approximately 2.2 Ma. These time estimates strongly suggest that C. yjhui is an ancient endemic of the Urugua-í basin and that it was present in the Urugua-í River basin for a long time, definitely much earlier than is the date of the construction of the hydroelectric dam (see Habitat above). The species, however, appears to have significantly increased in abundance only after the construction of the artificial lake (see Habitat).
Crenicichla yjhui shares the Urugua-í basin with two other closely related endemic Crenicichla species (C. ypo, C. yaha) and is separated from C. mandelburgeri by the 28 m high Urugua-í falls, which are located eight km from the confluence with the Paraná River (in 1989 the falls were replaced by a hydroelectric dam). The Urugua-í falls appear to have been a formidable barrier to upstream fish migration since the Urugua-í basin features a very high level of endemism (eight endemic species; Mirande & Koerber, 2015). Apart from the three endemic Crenicichla species there are also one endemic species of Gymnogeophagus and one of Australoheros among the cichlids plus several other endemics in other fish families. Based on the complex pattern of divergence C. yjhui diverged from the allopatric C. mandelburgeri (separated by the falls) between 0.41 Ma (mtDNA) and 0.85–2.2 Ma (nDNA; see above). The only other fish group where the divergence across the Urugua-í falls has been dated is the G. setequedas Reis, 1992 group where the Urugua-í endemic G. che diverged from its sister species between 1.39 and 0.76 Ma (Říčan et al., 2018). These ages derived from two cichlid genera are similar and fall within the Pleistocene epoch (see below).
Crenicichla mandelburgeri is in mtDNA markers not monophyletic (as opposed to nDNA) with C. yjhui found as the sister group of one of its two subclades. The two mtDNA clades of C. mandelburgeri are biogeographically delineated with a north–south division (Figs. 5, 6, 9). Crenicichla yjhui is the sister group of the northern mtDNA clade (i.e., of C. mandelburgeri sensu stricto) which makes good biogeographic sense since C. yjhui is located within the distributional area of this northern clade (Fig. 5). The reason for the N-S division in C. mandelburgeri can presently only be speculated upon but it is not the only species or species group (see e.g., the G. setequedas group in Říčan et al., 2018) with a similar biogeographic barrier within the same area of the Paraná valley without any presently evident physical barrier. Our hypothesis for the explanation of the divergence and the existence of these southern and northern clades is that it occurred during the Pleistocene epoch (2.58–0.012 Ma) when mean sea levels were at − 60 m and at minima at − 120 m (Haq et al., 1987) compared to today. This lowering of the sea level must have drastically enhanced isolation at existing barriers but most likely revealed additional barriers within the rio Paraná bed that are now submerged in the presently (interglacially) deep canyon of the rio Paraná in this middle section of the Paraná between Misiones, Argentina and Paraguay.
Conclusions
Ecological opportunity resulting from colonization of novel ecosystems is the driver of parallel adaptive radiations. Similar adaptive landscapes favor the evolution of similar suites of ecomorphs despite independent evolutionary histories. Here, we demonstrate that ecological opportunity has given rise to a rare (i.e., pelagic) adaptation in rivers and that the same adaptation has evolved in parallel in two cichlid species complexes in neighboring South American rivers. Recently, after the construction of an artificial lake, the pelagic adaptation has been seen to be very advantageous, permitting the species to significantly increase its abundance. Other sympatric species of the same species complex with a different set of adaptations (e.g., C. yaha), on the other hand, are due to the artificial lake facing lowered ecological opportunity because their habitat requirements are not supported there and the species is actually facing extinction in the whole river basin (Říčan et al., 2017). Invasive species now additionally threaten most endemic species in the Urugua-í with C. yjhui being so far one of the exceptions due to its adaptations, because most so-far introduced species are opportunistic omnivores. However, introduction of non-native predatory species might again lower the ecological opportunity and cause lowered fitness for C. yjhui.
Comparative material
A list of comparative material of C. scottii and C. vittata is available in Casciotta (1987). In addition, the following material was studied: All materials come from Argentina except as noted.
Crenicichla hu. MACN-ict 9429, holotype, 118.0 mm SL, Misiones, río Paraná basin, arroyo Piray–Miní. MACN-ict 9430, paratypes, 17 ex., 76.9–153.0 mm SL, same data as holotype.
Crenicichla iguassuensis. FMNH 54159, holotype, 137.0 mm SL, Brazil, Rio Iguaçu, Porto Uniăo da Victoria.
Crenicichla jupiaensis. MLP 11294, 2 ex., 87.7–93.0 mm SL, Corrientes, río Paraná at Yahapé.
Crenicichla lepidota Hensel, 1870. MACN-ict 7275, 1 ex., 151.6 mm SL, Corrientes, río Paraná basin, Isla Apipé Grande, Ituzaingó. MACN-ict 4091, 1 ex., 98.4 mm SL, Formosa, río Paraguay basin, Riacho de Oro. MACN-ict 3656, 2 ex., 116.0–165.7 mm SL, Misiones, Represa Estación Experimental Cerro Azul.
Crenicichla mandelburgeri. Misiones. MACN-ict 9442, 2 ex., 102.2–208.0 mm SL, Misiones, río Paraná basin, arroyo Cuńapirú, at route 223 near Ruiz de Montoya. MACN-ict 9440, 2 ex., 72.6–82.3 mm SL, Misiones, río paraná basin, arroyo Cuńapirú (arroyo Tucangua). MACN-ict 9441, 7 ex., 56.0–93.0 mm SL, Misiones, río Paraná basin, arroyo Guaruhape at route 220.
Crenicichla ocellata (Perugia, 1897). MSNG 33700, holotype, 257.5 mm SL, Paraguay, Puerto 14 de Mayo, Bahía Negra, Chaco Boreal.
Crenicichla semifasciata (Heckel, 1840). MACN-ict 6239, 1 ex., 176.6 mm SL, Formosa, río Paraguay basin, Riacho de Oro.
Crenicichla taikyra MACN-ict 9461, holotype, 98.3 mm SL, Misiones, río Paraná at Candelaria.Crenicichla tapii. Misiones. MLP 10560, holotype, 105.4 mm SL, Argentina, Misiones, río Iguazú basin, mouth at arroyo Ńandú.
Crenicichla tuca. Misiones. MLP 10818, holotype, 150.3 mm SL, río Iguazú basin, arroyo Deseado.
Crenicichla tesay MACN-ict 9016, holotype, 115.1 mm SL, Misiones, río Iguazú basin, arroyo Verde.
Crenicichla yaha. Misiones. MACN-ict 8924, holotype, 103.7 mm SL, río Paraná basin, arroyo Urugua-í at provincial route 19, Parque Provincial Islas Malvinas. MTD-F 30606, paratype, 1 ex., 105.9 mm SL, río Paraná basin, arroyo Urugua-í at provincial route 19, arroyo Uruzú, Parque Provincial Islas Malvinas. MLP 10909 (ex AI 200), paratype, 1 ex., 135.8 mm SL, río Paraná basin, arroyo Urugua-í at Isla Palacio.
Crenicichla ypo: Misiones. MACN-ict 9431, holotype, 105.5 mm SL, río Paraná basin, arroyo Urugua-í, at Establecimiento “Alto Paraná.” MACN-ict 9432, paratypes 3 ex., 101.0–116.0 mm SL, río Paraná basin, arroyo Urugua-í basin, arroyo Grapia, 6 km north from Colonia Gobernador J. J. Lanusse.
References
Alexander, D. H., J. Novembre & K. Lange, 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Research 19: 1655–1664.
Andrews, S., 2010. FastQC. A quality control tool for high throughput sequence data. [available on internet at http://www.bioinformatics.babraham.ac.uk/projects/fastqc].
Barlow, G. W., 2000. The Cichlid Fishes: Nature´s Grand Experiment in Evolution. Perseus Press, New York.
Burress, E. D., 2015. Cichlid fishes as models of ecological diversification: patterns, mechanisms, and consequences. Hydrobiologia 748: 7–27.
Burress, E. D., 2016. Ecological diversification associated with the pharyngeal jaw diversity of Neotropical cichlid fishes. Journal of Animal Ecology 85: 302–313.
Burress, E. D., A. Duarte, W. S. Serra, M. Loueiro, M. M. Gangloff & L. Siefferman, 2013. Functional diversification within a predatory species flock. PLoS ONE 8: e80929.
Burress, E. D., A. Duarte, W. S. Serra & M. Loureiro, 2015. Rates of piscivory predict pharyngeal jaw morphology in a piscivorous lineage of cichlid fishes. Ecology of Freshwater Fish 25: 590–598.
Burress, E. D., F. Alda, A. Duarte, M. Loureiro, J. W. Armbruster & P. Chakrabarty, 2018a. Phylogenomics of pike cichlids (Cichlidae: Crenicichla): the rapid ecological speciation of an incipient species flock. Journal of Evolutionary Biology 31: 14–30.
Burress, E. D., L. Piálek, J. Casciotta, A. Almirón, M. Tan, J. W. Armbruster & O. Říčan, 2018b. Island- and lake-like parallel adaptive radiations replicated in rivers. Proceedings of the Royal Society B 285: 20171762.
Casciotta, J. R., 1987. Crenicichla celidochilus n. sp. from Uruguay and a multivariate analysis of the lacustris group (Perciformes, Cichlidae). Copeia 1987: 883–891.
Casciotta, J. R. & G. Arratia, 1993. Jaws and teeth of American Cichlids (Pisces: Labroidei). Journal of Morphology 217: 1–36.
Casciotta, J., A. Almirón, L. Piálek, S. Gómez & O. Říčan, 2010. Crenicichla ypo (Teleostei: Cichlidae), a new species from the middle Paraná basin in Misiones, Argentina. Neotropical Ichthyology 8: 643–648.
Catchen, J. M., A. Amores, P. Hohenlohe, W. Cresko & J. H. Postlethwait, 2011. Stacks: building and genotyping loci de novo from short-read sequences. Bethesda 1: 171–182.
Chakrabarty, P., M. Warren, L. M. Page & C. C. Baldwin, 2013. GenSeq: an updated nomenclature and ranking for genetic sequences from type and non-type sources. Zookeys 346: 29–41.
Chifman, J. & L. Kubatko, 2014. Quartet-inference from SNP data under the coalescent model. Bioinformatics 30: 3317–3324.
de Queiroz, K., 2007. Species concepts and species delimitation. Systematic Biology 56: 879–886.
Drummond, A. J. & A. Rambaut, 2007. BEAST: bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 214.
Edgar, R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797.
Ferraris, C. J., 2007. Checklist of catfishes, recent and fossil (Osteichthyes: Siluriformes), and catalogue of siluriform primary types. Zootaxa 1418: 1–628.
Fryer, G. & T. D. Iles, 1972. The Cichlid fishes of the Great Lakes of Africa: their biology and evolution. T.F.H. Publications, New Jersey.
Givnish, T. J., 2015. Adaptive radiation versus ‘radiation’and ‘explosive diversification’: why conceptual distinctions are fundamental to understanding evolution. New Phytologist 207: 297–303.
Haq, B. U., J. Hardenbol & P. R. Vail, 1987. Chronology of fluctuating sea levels since the Triassic. Science 235: 1156–1167.
Jones, J. C., S. Fan, P. Franchini, M. Schartl & A. Meyer, 2013. The evolutionary history of Xiphophorus fish and their sexually selected sword: a genome-wide approach using restriction site-associated DNA sequencing. Molecular Ecology 22: 2986–3001.
Joyce, D. A., D. H. Lunt, R. Bills, G. F. Turner, C. Katongo, N. Duftner, C. Sturmbauer & O. Seehausen, 2005. An extant cichlid fish radiation emerged in an extinct Pleistocene lake. Nature 435: 90–95.
Kearse, M., R. Moir, A. Wilson, S. Stones-Havas, M. Cheung, S. Sturrock, S. Buxton, A. Cooper, S. Markowitz, C. Duran, T. Thierer, B. Ashton, P. Meintjes & A. Drummond, 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649.
Koblmüller, S., K. M. Sefc & C. Sturmbauer, 2008. The Lake Tanganyika cichlid species assemblage: recent advances in molecular phylogenetics. Hydrobiologia 615: 5–20.
Kullander, S. O., 1981. Cichlid fishes from the La Plata basin. Part I. Collections from Paraguay in the Muséum d’Histoire naturelle de Genève. Revue Suisse De Zoologie 88: 675–692.
Kullander, S. O., 1982. Cichlid fishes from the La Plata basin. Part III. The Crenicichla lepidota species group. Revue Suisse De Zoologie 89: 627–661.
Kullander, S. O., 1986. Cichlid fishes of the Amazon River drainage of Peru. Swedish Museum of Natural History, Stockholm.
Kullander, S. O., 1996. Heroina isonycterina, a new genus and species of cichlid fish from Western Amazonia, with comments on cichlasomine sys-tematics. Ichthyological Exploration of Freshwaters 7: 149–172.
Kullander, S. O., 2009. Crenicichla mandelburgeri, a new species of cichlid fish (Teleostei: Cichlidae) from the Paraná river drainage in Paraguay. Zootaxa 2006: 41–50.
Langmead, B. & S. L. Salzberg, 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9: 357–359.
Lucena, C. A. S. & S. O. Kullander, 1992. The Crenicichla (Teleostei: Cichlidae) species of the Uruguai River drainage in Brazil. Ichthyological Exploration of Freshwaters 3: 97–160.
Matschiner, M., Z. Musilová, J. M. Barth, Z. Starostová, W. Salzburger, M. Steel & R. Bouckaert, 2017. Bayesian phylogenetic estimation of clade ages supports trans-atlantic dispersal of cichlid fishes. Systematic Biology 66: 3–22.
Mirande, J. M. & S. Koerber, 2015. Checklist of the freshwater fishes of argentina (CLOFFAR). Ichthyological Contributions of PecesCriollos 36: 1–68.
Musilová, Z., O. Říčan, Š. Říčanová, P. Janšta, O. Gahura & J. Novák, 2015. Phylogeny and historical biogeography of trans-Andean cichlid fishes (Teleostei: Cichlidae). Vertebrate Zoology 65: 333–350.
Peterson, B. K., J. N. Weber, E. H. Kay, H. S. Fisher & H. E. Hoekstra, 2012. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7: e37135.
Piálek, L., O. Říčan, J. Casciotta, A. Almirón & J. Zrzavý, 2012. Multilocus phylogeny of Crenicichla (Teleostei: Cichlidae), with biogeography of the C. lacustris group: species flocks as a model for sympatric speciation in rivers. Molecular Phylogenetics and Evolution 62: 46–61.
Piálek, L., K. Dragová, J. Casciotta, A. Almirón & O. Říčan, 2015. Description of two new species of Crenicichla (Teleostei: Cichlidae) from the lower Iguazú River with a taxonomic reappraisal of C. iguassuensis, C. tesay and C. yaha. Historia Natural Tercera Serie 5: 5–27.
Piálek, L., E. Burress, K. Dragová, A. Almirón, J. Casciotta & O. Říčan, 2018. Phylogenomics of pike cichlids (Cichlidae: Crenicichla) of the C. mandelburgeri species complex: rapid ecological speciation in the Iguazú River and high endemism in the Middle Paraná basin. Hydrobiologia in Press.
Posada, D., 2008. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256.
Purcell, S., B. Neale, K. Todd-Brown, L. Thomas, M. A. Ferreira, D. Bender, J. Maller, P. Sklar, P. I. de Bakker, M. J. Daly & P. C. Sham, 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics 81: 559–575.
Rambaut, A., M. A. Suchard & A. J. Drummond, 2014. Tracer v1.6. [available on internet at http://tree.bio.ed.ac.uk/software/tracer].
Říčan, O., L. Piálek, K. Dragová & J. Novák, 2016. Diversity and evolution of the Middle American cichlid fishes (Teleostei: Cichlidae) with revised classification. Vertebrate Zoology 66: 1–102.
Říčan, O., A. Almirón & J. Casciotta, 2017. Rediscovery of Crenicichla yaha (Teleostei: Cichlidae). Ichthyological Contributions of PecesCriollos 50: 1–8.
Říčan, O., Š. Říčanová, K. Dragová, L. Piálek, A. Almirón & J. Casciotta, 2018. Species diversity in Gymnogeophagus (Teleostei: Cichlidae) and comparative biogeography of cichlids in the Middle Paraná basin, an emerging hotspot of fish endemism. Hydrobiologia in Press. https://doi.org/10.1007/s10750-018-3691-z.
Schluter, D., 2000. The Ecology of Adaptive Radiation. OUP, Oxford.
Seehausen, O., 2015. Process and pattern in cichlid radiations–inferences for understanding unusually high rates of evolutionary diversification. New Phytologist 207: 304–312.
Seehausen, O., P. J. Mayhew & J. J. M. Van Alphen, 1999. Evolution of colour patterns in East African cichlid fish. Journal of Evolutionary Biology 12: 514–534.
Simpson, G. G., 1953. The Major Features of Evolution. Columbia University Press, New York.
Šmilauer, P. & J. Lepš, 2014. Multivariate Analysis of Ecological Data using Canoco 5. Cambridge University Press, New York.
Stamatakis, A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313.
Swofford, D. L., 2003. PAUP*. Phylogenetic analysis using parsimony (* and other methods). Version 4 [Computer Programme]. Sinauer Associates, Sunderland, MA.
Takahashi, T. & E. Moreno, 2015. A RAD-based phylogenetics for Orestias fishes from Lake Titicaca. Molecular Phylogenetics and Evolution 93: 307–317.
Takahashi, T., N. Nagata & T. Sota, 2014. Application of RAD-based phylogenetics to complex relationships among variously related taxa in a species flock. Molecular Phylogenetics and Evolution 80: 137–144.
Taylor, W. R. & G. C. Van Dyke, 1985. Revised procedures for staining and clearing small fishes and other vertebrates for bone and cartilage study. Cybium 9: 107–119.
Acknowledgements
The authors thank Soledad Gouric for the pictures of the LPJ and the authorities of Ministerio de Ecología y Recursos Naturales Renovables de la Provincia de Misiones for awarding fishing permits. We are very grateful to Vladimír Beneš, Bianka Baying, and the EMBL Genomic Core Facility in Heidelberg (Germany) for their kind advice and technical support during the DNA library finalization and Illumina sequencing. Financial support was provided by the Czech Science Foundation (GAČR) under Grant Number 14-28518P to LP; by Comisión de Investigaciones Científicas de la provincia de Buenos Aires (CIC); Facultad de Ciencias Naturales y Museo (UNLP); and Administración de Parques Nacionales. The access to computing and storage facilities owned by parties and projects contributing to the National Grid Infrastructure MetaCentrum provided under the program “Projects of Large Infrastructure for Research, Development, and Innovations” (LM2010005) was highly appreciated, as well as the access to the CERIT-SC computing and storage facilities provided under the program Center CERIT Scientific Cloud, part of the Operational Program Research and Development for Innovations, Reg. No. CZ. 1.05/3.2.00/08.0144.
Author information
Authors and Affiliations
Corresponding author
Additional information
Guest editors: S. Koblmüller, R. C. Albertson, M. J. Genner, K. M. Sefc & T. Takahashi / Advances in Cichlid Research III: Behavior, Ecology and Evolutionary Biology
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1.
Localities of C. yjhui. A) type locality, Urugua-í reservoir at camping, Misiones, Argentina (25°52′29.1″S–54°33′02.2″W). B) paratype locality, Urugua-í reservoir opposite Policia Fluvial Station (25°54′02.2″S, 54°33′18.6″W); C) paratype locality, arroyo Falso Urugua-í (25°58′26″S, 54°15′29″W). Supplementary material 1 (JPEG 2549 kb)
Table S1.
Specimens included in the study. Supplementary material 2 (XLSX 38 kb)
Table S2.
Summary of SVDQ analyses under different parameter settings with focus on relationships of C. yjhui. We have analyzed matrices with 50% and 70% SNPs, with Stacks depth of 3 and 8, and with inclusion of homozygotic, homozygotic + heterozygotic, 1st only SNP homozygotic + heterozygotic and all. The placement of sister-species relationship either with C. mandelburgeri or C. ypo is randomly distributed across the matrices. Note that in all cases support for placement with the respective species (either with C. mandelburgeri or C. ypo) is 100% in bootstrap analyses. Supplementary material 3 (XLS 15 kb)
Rights and permissions
About this article
Cite this article
Piálek, L., Casciotta, J., Almirón, A. et al. A new pelagic predatory pike cichlid (Teleostei: Cichlidae: Crenicichla) from the C. mandelburgeri species complex with parallel and reticulate evolution. Hydrobiologia 832, 377–395 (2019). https://doi.org/10.1007/s10750-018-3754-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-018-3754-1